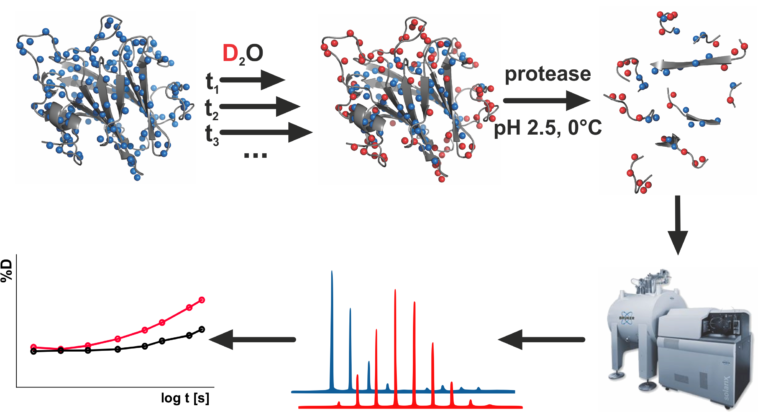
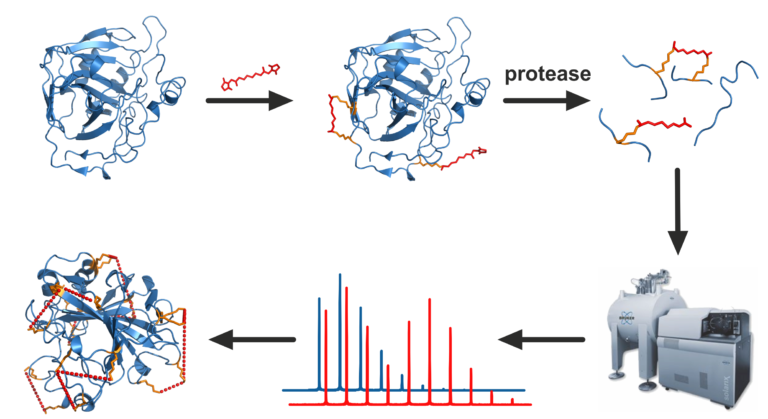
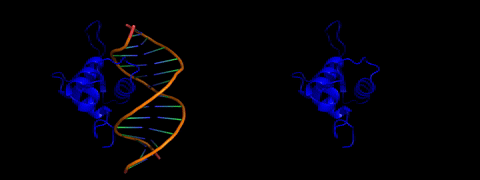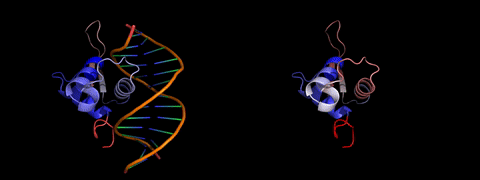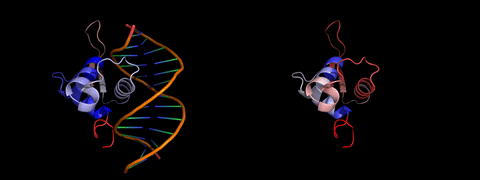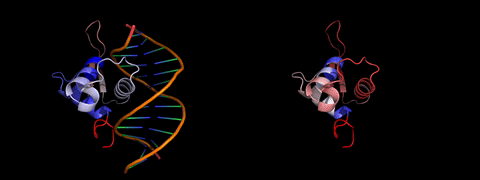
About laboratory
Laboratory of Structural Biology and Cell Signaling consists of a multidisciplinary team whose thematic focus is divided into two main directions. Structural biology group produces proteins by recombinant expression technologies and characterizes the protein or protein-ligand structures and their dynamics utilizing advanced mass spectrometric techniques. Cell signaling unit examines the linkages of cellular signaling to the metabolism of cancer cells using molecular biology and biochemistry methods. Through the collaboration of the two groups a unique research platform is formed, wherein the results obtained by the study of biological systems can also be verified and explained at the molecular level. Laboratory is located at the BIOCEV center in Vestec.